import pandas as pd
# TB incidence in Africa
tb_2019_africa = pd.read_csv(
"https://raw.githubusercontent.com/the-graph-courses/idap_book/main/data/tb_incidence_2019.csv"
)
# Health expenditure data
health_exp_2019 = pd.read_csv(
"https://raw.githubusercontent.com/the-graph-courses/idap_book/main/data/health_expend_per_cap_2019.csv"
)
# Highest expenditure countries
highest_exp = health_exp_2019.sort_values("expend_usd", ascending=False).head(70)
# TB cases in children
tb_cases_children = pd.read_csv(
"https://raw.githubusercontent.com/the-graph-courses/idap_book/main/data/tb_cases_children_2012.csv"
).dropna()
# Country continents data
country_continents = pd.read_csv(
"https://raw.githubusercontent.com/the-graph-courses/idap_book/main/data/country_continents.csv"
)
# people data
people = pd.DataFrame({"name": ["Alice", "Bob", "Charlie"], "age": [25, 32, 45]})
# Test information
test_info = pd.DataFrame(
{
"name": ["Alice", "Bob", "Charlie"],
"test_date": ["2023-06-05", "2023-08-10", "2023-07-15"],
"result": ["Negative", "Positive", "Negative"],
}
)
# Disordered test information
test_info_disordered = pd.DataFrame(
{
"name": ["Bob", "Alice", "Charlie"], # Bob in first row
"test_date": ["2023-08-10", "2023-06-05", "2023-07-15"],
"result": ["Positive", "Negative", "Negative"],
}
)
# Multiple test information
test_info_multiple = pd.DataFrame(
{
"name": ["Alice", "Alice", "Bob", "Charlie"],
"test_date": ["2023-06-05", "2023-06-06", "2023-08-10", "2023-07-15"],
"result": ["Negative", "Negative", "Positive", "Negative"],
}
)
# Test information with different name
test_info_different_name = pd.DataFrame(
{
"first_name": ["Alice", "Bob", "Charlie"],
"test_date": ["2023-06-05", "2023-08-10", "2023-07-15"],
"result": ["Negative", "Positive", "Negative"],
}
)
# Test information including Xavier
test_info_xavier = pd.DataFrame(
{
"name": ["Alice", "Bob", "Xavier"],
"test_date": ["2023-06-05", "2023-08-10", "2023-05-02"],
"result": ["Negative", "Positive", "Negative"],
}
)
# Students data
students = pd.DataFrame(
{"student_id": [1, 2, 3], "name": ["Alice", "Bob", "Charlie"], "age": [20, 22, 21]}
)
# Exam dates data
exam_dates = pd.DataFrame(
{"student_id": [1, 3], "exam_date": ["2023-05-20", "2023-05-22"]}
)
# Employee details
employee_details = pd.DataFrame(
{
"id_number": ["E001", "E002", "E003"],
"full_name": ["Emily", "Frank", "Grace"],
"department": ["HR", "IT", "Marketing"],
}
)
# Performance reviews
performance_reviews = pd.DataFrame(
{
"employee_code": ["E001", "E002", "E003"],
"review_type": ["Annual", "Mid-year", "Annual"],
"review_date": ["2022-05-10", "2023-09-01", "2021-12-15"],
}
)
# Sales data
sales_data = pd.DataFrame(
{
"salesperson_id": [1, 4, 8],
"product": ["Laptop", "Smartphone", "Tablet"],
"date_of_sale": ["2023-01-15", "2023-03-05", "2023-02-20"],
}
)
# Salesperson peoples
salesperson_peoples = pd.DataFrame(
{
"salesperson_id": [1, 2, 3, 5, 8],
"name": ["Alice", "Bob", "Charlie", "Diana", "Eve"],
"age": [28, 45, 32, 55, 40],
"gender": ["Female", "Male", "Male", "Female", "Female"],
}
)
# Total sales data
total_sales = pd.DataFrame(
{
"product": [
"Laptop",
"Desktop",
"Tablet",
"Smartphone",
"Smartwatch",
"Headphones",
"Monitor",
"Keyboard",
"Mouse",
"Printer",
],
"total_units_sold": [9751, 136, 8285, 2478, 3642, 5231, 1892, 4267, 3891, 982],
}
)
# Product feedback data
product_feedback = pd.DataFrame(
{
"product": [
"Laptop",
"Desktop",
"Tablet",
"Smartphone",
"Smartwatch",
"Headphones",
"Monitor",
"Gaming Console",
"Camera",
"Speaker",
],
"n_positive_reviews": [1938, 128, 842, 1567, 723, 956, 445, 582, 234, 678],
"n_negative_reviews": [42, 30, 56, 89, 34, 28, 15, 11, 8, 25],
}
)
# Sales incidence data
sales = pd.DataFrame(
{
"year": [2010, 2011, 2014, 2016, 2017],
"sales_count": [69890, 66507, 59831, 58704, 59151],
}
)
# Customer complaints data
customer_complaints = pd.DataFrame(
{
"year": [2011, 2013, 2015, 2016, 2019],
"complaints_count": [1292, 1100, 1011, 940, 895],
}
)
employees = pd.DataFrame(
{"employee_id": [1, 2, 3], "name": ["John", "Joy", "Khan"], "age": [32, 28, 40]}
)
training_sessions = pd.DataFrame(
{
"employee_id": [1, 2, 3],
"training_date": ["2023-01-20", "2023-02-20", "2023-05-15"],
}
)
customer_details = pd.DataFrame(
{
"id_number": ["A001", "B002", "C003"],
"full_name": ["Alice", "Bob", "Charlie"],
"address": ["123 Elm St", "456 Maple Dr", "789 Oak Blvd"],
}
)
# Order Records
order_records = pd.DataFrame(
{
"customer_code": ["A001", "B002", "C003"],
"product_type": ["Electronics", "Books", "Clothing"],
"order_date": ["2022-05-10", "2023-09-01", "2021-12-15"],
}
)21 Introduction to Joining Datasets
21.1 Data & Packages
Please run the code below to load the packages and datasets we’ll be using throughout this lesson.
21.2 Intro
Joining is a key skill when working with data as it allows you to combine information about the same entities from multiple sources, leading to more comprehensive and insightful analyses. In this lesson, you’ll learn how to use different joining techniques using Python’s pandas library. Let’s get started!
21.3 Learning Objectives
You understand how each of the different joins work: left, right, inner, and outer.
You can join simple datasets together using the
pd.merge()function.
21.4 Why Do We Need Joins?
To illustrate the utility of joins, let’s start with a toy example. Consider the following two datasets. The first, people, contains names and ages of three individuals:
people| name | age | |
|---|---|---|
| 0 | Alice | 25 |
| 1 | Bob | 32 |
| 2 | Charlie | 45 |
The second, test_info, contains test dates and results for those individuals:
test_info| name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 2023-08-10 | Positive |
| 2 | Charlie | 2023-07-15 | Negative |
We’d like to analyze these data together, and so we need a way to combine them.
One option we might consider is concatenating the dataframes horizontally using pd.concat():
pd.concat([people, test_info], axis=1)| name | age | name | test_date | result | |
|---|---|---|---|---|---|
| 0 | Alice | 25 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 32 | Bob | 2023-08-10 | Positive |
| 2 | Charlie | 45 | Charlie | 2023-07-15 | Negative |
This successfully merges the datasets, but it doesn’t do so very intelligently. The function essentially “pastes” or “staples” the two tables together. So, as you can notice, the “name” column appears twice. This is not ideal and will be problematic for analysis.
Another problem occurs if the rows in the two datasets are not already aligned. In this case, the data will be combined incorrectly with pd.concat(). Consider the test_info_disordered dataset, which now has Bob in the first row:
test_info_disordered| name | test_date | result | |
|---|---|---|---|
| 0 | Bob | 2023-08-10 | Positive |
| 1 | Alice | 2023-06-05 | Negative |
| 2 | Charlie | 2023-07-15 | Negative |
What happens if we concatenate this with the original people dataset, where Bob was in the second row?
pd.concat([people, test_info_disordered], axis=1)| name | age | name | test_date | result | |
|---|---|---|---|---|---|
| 0 | Alice | 25 | Bob | 2023-08-10 | Positive |
| 1 | Bob | 32 | Alice | 2023-06-05 | Negative |
| 2 | Charlie | 45 | Charlie | 2023-07-15 | Negative |
Alice’s people details are now mistakenly aligned with Bob’s test info!
A third issue arises when an entity appears more than once in one dataset. Perhaps Alice had multiple tests:
test_info_multiple| name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Alice | 2023-06-06 | Negative |
| 2 | Bob | 2023-08-10 | Positive |
| 3 | Charlie | 2023-07-15 | Negative |
If we try to concatenate this with the people dataset, we’ll get mismatched data due to differing row counts:
pd.concat([people, test_info_multiple], axis=1)| name | age | name | test_date | result | |
|---|---|---|---|---|---|
| 0 | Alice | 25.0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 32.0 | Alice | 2023-06-06 | Negative |
| 2 | Charlie | 45.0 | Bob | 2023-08-10 | Positive |
| 3 | NaN | NaN | Charlie | 2023-07-15 | Negative |
This results in NaN values and misaligned data.
What we have here is called a one-to-many relationship—one Alice in the people data, but multiple Alice rows in the test data since she had multiple tests. Joining in such cases will be covered in detail in the second joining lesson.
Clearly, we need a smarter way to combine datasets than concatenation; we’ll need to venture into the world of joining. In pandas, the function that performs joins is pd.merge().
It works for the simple case, and it does not duplicate the name column:
pd.merge(people, test_info)| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
| 2 | Charlie | 45 | 2023-07-15 | Negative |
It works where the datasets are not ordered identically:
pd.merge(people, test_info_disordered)| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
| 2 | Charlie | 45 | 2023-07-15 | Negative |
As you can see, Alice’s details are now correctly aligned with her test results.
And it works when there are multiple test rows per individual:
pd.merge(people, test_info_multiple)| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Alice | 25 | 2023-06-06 | Negative |
| 2 | Bob | 32 | 2023-08-10 | Positive |
| 3 | Charlie | 45 | 2023-07-15 | Negative |
In this case, the pd.merge() function correctly repeats Alice’s details for each of her tests.
Simple and beautiful!
21.5 pd.merge() syntax
Now that we understand why we need joins, let’s look at their basic syntax.
Joins take two dataframes as the first two arguments: left (the left dataframe) and right (the right dataframe). In pandas, you can provide these as positional or keyword arguments:
# left and right
pd.merge(left=people, right=test_info) # keyword arguments
pd.merge(people, test_info) # positional arguments| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
| 2 | Charlie | 45 | 2023-07-15 | Negative |
Another critical argument is on, which indicates the column or key used to connect the tables. We don’t always need to supply this argument; it can be inferred from the datasets. For example, in our original examples, “name” is the only column common to people and test_info. So the merge function assumes on='name':
# on argument is optional if the column key is the same in both dataframes
pd.merge(people, test_info)
pd.merge(people, test_info, on="name")| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
| 2 | Charlie | 45 | 2023-07-15 | Negative |
The column used to connect rows across the tables is known as a key. In the pandas merge() function, the key is specified in the on argument, as seen in pd.merge(people, test_info, on='name').
What happens if the keys are named differently in the two datasets? Consider the test_info_different_name dataset, where the “name” column has been changed to “first_name”:
test_info_different_name| first_name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 2023-08-10 | Positive |
| 2 | Charlie | 2023-07-15 | Negative |
If we try to join test_info_different_name with our original people dataset, we will encounter an error:
pd.merge(people, test_info_different_name)MergeError: No common columns to perform merge on. Merge options: left_on=None, right_on=None, left_index=False, right_index=FalseThe error indicates that there are no common variables, so the join is not possible.
In situations like this, you have two choices: you can rename the column in the second dataframe to match the first, or more simply, specify which columns to join on using left_on and right_on.
Here’s how to do this:
pd.merge(people, test_info_different_name, left_on='name', right_on='first_name')| name | age | first_name | test_date | result | |
|---|---|---|---|---|---|
| 0 | Alice | 25 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 32 | Bob | 2023-08-10 | Positive |
| 2 | Charlie | 45 | Charlie | 2023-07-15 | Negative |
This syntax essentially says, “Connect name from the left dataframe with first_name from the right dataframe because they represent the same data.”
Key: The column or set of columns used to match rows between two dataframes in a join operation.
Left Join: A type of join that keeps all rows from the left dataframe and adds matching rows from the right dataframe. If there is no match, the result is NaN on the right side.
21.6 Practice Q: Join Employees and Training Sessions
Consider the two datasets below, one with employee details and the other with training session dates for these employees.
employees| employee_id | name | age | |
|---|---|---|---|
| 0 | 1 | John | 32 |
| 1 | 2 | Joy | 28 |
| 2 | 3 | Khan | 40 |
training_sessions| employee_id | training_date | |
|---|---|---|
| 0 | 1 | 2023-01-20 |
| 1 | 2 | 2023-02-20 |
| 2 | 3 | 2023-05-15 |
How many rows and columns would you expect to have after joining these two datasets?
Now join the two datasets and check your answer.
# Your code here21.7 Practice Q: Join with on Argument
Two datasets are shown below, one with customer details and the other with order records for those customers.
customer_details| id_number | full_name | address | |
|---|---|---|---|
| 0 | A001 | Alice | 123 Elm St |
| 1 | B002 | Bob | 456 Maple Dr |
| 2 | C003 | Charlie | 789 Oak Blvd |
order_records| customer_code | product_type | order_date | |
|---|---|---|---|
| 0 | A001 | Electronics | 2022-05-10 |
| 1 | B002 | Books | 2023-09-01 |
| 2 | C003 | Clothing | 2021-12-15 |
Join the customer_details and order_records datasets. You will need to use the left_on and right_on arguments because the customer identifier columns have different names.
21.8 Types of joins
The toy examples so far have involved datasets that could be matched perfectly—every row in one dataset had a corresponding row in the other dataset.
Real-world data is usually messier. Often, there will be entries in the first table that do not have corresponding entries in the second table, and vice versa.
To handle these cases of imperfect matching, there are different join types with specific behaviors: left, right, inner, and outer. In the upcoming sections, we’ll look at examples of how each join type operates on datasets with imperfect matches.
21.9 left join
Let’s start with the left join. To see how it handles unmatched rows, we will try to join our original people dataset with a modified version of the test_info dataset.
As a reminder, here is the people dataset, with Alice, Bob, and Charlie:
people| name | age | |
|---|---|---|
| 0 | Alice | 25 |
| 1 | Bob | 32 |
| 2 | Charlie | 45 |
For test information, we’ll remove Charlie and we’ll add a new individual, Xavier, and his test data:
test_info_xavier| name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 2023-08-10 | Positive |
| 2 | Xavier | 2023-05-02 | Negative |
We can specify the join type using the how argument:
pd.merge(people, test_info_xavier, how='left')| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
| 2 | Charlie | 45 | NaN | NaN |
As you can see, with the left join, all records from the left dataframe (people) are retained. So, even though Charlie doesn’t have a match in the test_info_xavier dataset, he’s still included in the output. (But of course, since his test information is not available in test_info_xavier, those values were left as NaN.)
Xavier, on the other hand, who was only present in the right dataset, gets dropped.
The graphic below shows how this join worked:

Now what if we flip the dataframes? Let’s see the outcome when test_info_xavier is the left dataframe and people is the right one:
pd.merge(test_info_xavier, people, on='name', how='left')| name | test_date | result | age | |
|---|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative | 25.0 |
| 1 | Bob | 2023-08-10 | Positive | 32.0 |
| 2 | Xavier | 2023-05-02 | Negative | NaN |
Once again, the left join retains all rows from the left dataframe (now test_info_xavier). This means Xavier’s data is included this time. Charlie, on the other hand, is excluded.
Primary Dataset: In the context of joins, the primary dataset refers to the main or prioritized dataset in an operation. In a left join, the left dataframe is considered the primary dataset because all of its rows are retained in the output, regardless of whether they have a matching row in the other dataframe.
21.10 Practice Q: Left Join Students and Exam Dates
Consider the two datasets below, one with student details and the other with exam dates for some of these students.
students| student_id | name | age | |
|---|---|---|---|
| 0 | 1 | Alice | 20 |
| 1 | 2 | Bob | 22 |
| 2 | 3 | Charlie | 21 |
exam_dates| student_id | exam_date | |
|---|---|---|
| 0 | 1 | 2023-05-20 |
| 1 | 3 | 2023-05-22 |
Join the students dataset with the exam_dates dataset using a left join.
21.11 Analysing African TB Incidence and Health Expenditure
Let’s try another example, this time with a more realistic set of data.
First, we have data on the TB incidence rate per 100,000 people for some African countries, from the WHO:
tb_2019_africa| country | cases | conf_int_95 | |
|---|---|---|---|
| 0 | Burundi | 107 | [69 – 153] |
| 1 | Sao Tome and Principe | 114 | [45 – 214] |
| 2 | Senegal | 117 | [83 – 156] |
| 3 | Mauritius | 12 | [9 – 15] |
| 4 | Côte d’Ivoire | 137 | [88 – 197] |
| 5 | Ethiopia | 140 | [98 – 188] |
| 6 | Chad | 142 | [92 – 202] |
| 7 | Ghana | 144 | [70 – 244] |
| 8 | Malawi | 146 | [78 – 235] |
| 9 | Seychelles | 15 | [13 – 18] |
| 10 | Gambia | 158 | [117 – 204] |
| 11 | Guinea | 176 | [114 – 251] |
| 12 | Cameroon | 179 | [116 – 255] |
| 13 | Zimbabwe | 199 | [147 – 258] |
| 14 | Uganda | 200 | [117 – 303] |
| 15 | Nigeria | 219 | [143 – 311] |
| 16 | South Sudan | 227 | [147 – 324] |
| 17 | Madagascar | 233 | [151 – 333] |
| 18 | United Republic of Tanzania | 237 | [112 – 408] |
| 19 | Botswana | 253 | [195 – 317] |
| 20 | Kenya | 267 | [163 – 396] |
| 21 | Equatorial Guinea | 286 | [185 – 408] |
| 22 | Sierra Leone | 295 | [190 – 422] |
| 23 | Liberia | 308 | [199 – 440] |
| 24 | Democratic Republic of the Congo | 320 | [207 – 457] |
| 25 | Zambia | 333 | [216 – 474] |
| 26 | Comoros | 35 | [23 – 50] |
| 27 | Angola | 351 | [227 – 501] |
| 28 | Mozambique | 361 | [223 – 532] |
| 29 | Guinea-Bissau | 361 | [234 – 516] |
| 30 | Eswatini | 363 | [228 – 527] |
| 31 | Togo | 37 | [30 – 45] |
| 32 | Congo | 373 | [237 – 541] |
| 33 | Cabo Verde | 46 | [35 – 58] |
| 34 | Burkina Faso | 47 | [30 – 67] |
| 35 | Namibia | 486 | [348 – 647] |
| 36 | Mali | 52 | [34 – 74] |
| 37 | Gabon | 521 | [337 – 744] |
| 38 | Central African Republic | 540 | [349 – 771] |
| 39 | Benin | 55 | [36 – 79] |
| 40 | Rwanda | 57 | [44 – 72] |
| 41 | Algeria | 61 | [46 – 77] |
| 42 | South Africa | 615 | [427 – 835] |
| 43 | Lesotho | 654 | [406 – 959] |
| 44 | Niger | 84 | [54 – 120] |
| 45 | Eritrea | 86 | [40 – 151] |
| 46 | Mauritania | 89 | [58 – 127] |
We want to analyze how TB incidence in African countries varies with government health expenditure per capita. For this, we have data on health expenditure per capita in USD, also from the WHO, for countries from all continents:
health_exp_2019| country | expend_usd | |
|---|---|---|
| 0 | Nigeria | 10.97 |
| 1 | Bahamas | 1002.00 |
| 2 | United Arab Emirates | 1015.00 |
| 3 | Nauru | 1038.00 |
| 4 | Slovakia | 1058.00 |
| ... | ... | ... |
| 180 | Myanmar | 9.64 |
| 181 | Malawi | 9.78 |
| 182 | Cuba | 901.80 |
| 183 | Tunisia | 97.75 |
| 184 | Nicaragua | 99.73 |
185 rows × 2 columns
Which dataset should we use as the left dataframe for the join?
Since our goal is to analyze African countries, we should use tb_2019_africa as the left dataframe. This will ensure we keep all the African countries in the final joined dataset.
Let’s join them:
tb_health_exp_joined = pd.merge(tb_2019_africa, health_exp_2019, on='country', how='left')
tb_health_exp_joined| country | cases | conf_int_95 | expend_usd | |
|---|---|---|---|---|
| 0 | Burundi | 107 | [69 – 153] | 6.07 |
| 1 | Sao Tome and Principe | 114 | [45 – 214] | 47.64 |
| 2 | Senegal | 117 | [83 – 156] | 15.47 |
| 3 | Mauritius | 12 | [9 – 15] | NaN |
| 4 | Côte d’Ivoire | 137 | [88 – 197] | 22.25 |
| 5 | Ethiopia | 140 | [98 – 188] | 5.93 |
| 6 | Chad | 142 | [92 – 202] | 4.76 |
| 7 | Ghana | 144 | [70 – 244] | 30.01 |
| 8 | Malawi | 146 | [78 – 235] | 9.78 |
| 9 | Seychelles | 15 | [13 – 18] | 572.00 |
| 10 | Gambia | 158 | [117 – 204] | 9.40 |
| 11 | Guinea | 176 | [114 – 251] | 9.61 |
| 12 | Cameroon | 179 | [116 – 255] | 6.26 |
| 13 | Zimbabwe | 199 | [147 – 258] | 7.82 |
| 14 | Uganda | 200 | [117 – 303] | 5.05 |
| 15 | Nigeria | 219 | [143 – 311] | 10.97 |
| 16 | South Sudan | 227 | [147 – 324] | NaN |
| 17 | Madagascar | 233 | [151 – 333] | 6.26 |
| 18 | United Republic of Tanzania | 237 | [112 – 408] | 16.02 |
| 19 | Botswana | 253 | [195 – 317] | 292.10 |
| 20 | Kenya | 267 | [163 – 396] | 39.57 |
| 21 | Equatorial Guinea | 286 | [185 – 408] | 47.30 |
| 22 | Sierra Leone | 295 | [190 – 422] | 6.28 |
| 23 | Liberia | 308 | [199 – 440] | 8.38 |
| 24 | Democratic Republic of the Congo | 320 | [207 – 457] | 3.14 |
| 25 | Zambia | 333 | [216 – 474] | 27.09 |
| 26 | Comoros | 35 | [23 – 50] | NaN |
| 27 | Angola | 351 | [227 – 501] | 28.59 |
| 28 | Mozambique | 361 | [223 – 532] | 9.35 |
| 29 | Guinea-Bissau | 361 | [234 – 516] | 3.90 |
| 30 | Eswatini | 363 | [228 – 527] | 131.50 |
| 31 | Togo | 37 | [30 – 45] | 7.56 |
| 32 | Congo | 373 | [237 – 541] | 25.82 |
| 33 | Cabo Verde | 46 | [35 – 58] | 111.50 |
| 34 | Burkina Faso | 47 | [30 – 67] | 17.17 |
| 35 | Namibia | 486 | [348 – 647] | 204.30 |
| 36 | Mali | 52 | [34 – 74] | 11.03 |
| 37 | Gabon | 521 | [337 – 744] | 125.60 |
| 38 | Central African Republic | 540 | [349 – 771] | 3.58 |
| 39 | Benin | 55 | [36 – 79] | 6.33 |
| 40 | Rwanda | 57 | [44 – 72] | 20.20 |
| 41 | Algeria | 61 | [46 – 77] | 163.00 |
| 42 | South Africa | 615 | [427 – 835] | 321.70 |
| 43 | Lesotho | 654 | [406 – 959] | 53.02 |
| 44 | Niger | 84 | [54 – 120] | 11.14 |
| 45 | Eritrea | 86 | [40 – 151] | 4.45 |
| 46 | Mauritania | 89 | [58 – 127] | 22.40 |
Now in the joined dataset, we have just the African countries, which is exactly what we wanted.
All rows from the left dataframe tb_2019_africa were kept, while non-African countries from health_exp_2019 were discarded.
We can check if any rows in tb_2019_africa did not have a match in health_exp_2019 by filtering for NaN values:
tb_health_exp_joined.query("expend_usd.isna()")| country | cases | conf_int_95 | expend_usd | |
|---|---|---|---|---|
| 3 | Mauritius | 12 | [9 – 15] | NaN |
| 16 | South Sudan | 227 | [147 – 324] | NaN |
| 26 | Comoros | 35 | [23 – 50] | NaN |
This shows that 3 countries—Mauritius, South Sudan, and Comoros—did not have expenditure data in health_exp_2019. But because they were present in tb_2019_africa, and that was the left dataframe, they were still included in the joined data.
21.12 Practice Q: Left Join TB Cases and Continents
The first, tb_cases_children, contains the number of TB cases in under 15s in 2012, by country:
tb_cases_children| country | tb_cases_smear_0_14 | |
|---|---|---|
| 0 | Afghanistan | 588.0 |
| 1 | Albania | 0.0 |
| 2 | Algeria | 89.0 |
| 4 | Andorra | 0.0 |
| 5 | Angola | 982.0 |
| ... | ... | ... |
| 211 | Viet Nam | 142.0 |
| 213 | West Bank and Gaza Strip | 0.0 |
| 214 | Yemen | 105.0 |
| 215 | Zambia | 321.0 |
| 216 | Zimbabwe | 293.0 |
200 rows × 2 columns
And country_continents, lists all countries and their corresponding region and continent:
country_continents| country.name.en | continent | region | |
|---|---|---|---|
| 0 | Afghanistan | Asia | South Asia |
| 1 | Albania | Europe | Europe & Central Asia |
| 2 | Algeria | Africa | Middle East & North Africa |
| 3 | American Samoa | Oceania | East Asia & Pacific |
| 4 | Andorra | Europe | Europe & Central Asia |
| ... | ... | ... | ... |
| 286 | Yugoslavia | NaN | Europe & Central Asia |
| 287 | Zambia | Africa | Sub-Saharan Africa |
| 288 | Zanzibar | NaN | Sub-Saharan Africa |
| 289 | Zimbabwe | Africa | Sub-Saharan Africa |
| 290 | Åland Islands | Europe | Europe & Central Asia |
291 rows × 3 columns
Your goal is to add the continent and region data to the TB cases dataset.
Which dataframe should be the left one? And which should be the right one? Once you’ve decided, join the datasets appropriately using a left join.
21.13 right join
A right join can be thought of as a mirror image of a left join. The mechanics are the same, but now all rows from the right dataframe are retained, while only those rows from the left dataframe that find a match in the right are kept.
Let’s look at an example to understand this. We’ll use our original people and modified test_info_xavier datasets:
people
test_info_xavier| name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 2023-08-10 | Positive |
| 2 | Xavier | 2023-05-02 | Negative |
Now let’s try a right join, with people as the right dataframe:
pd.merge(test_info_xavier, people, on='name', how='right')| name | test_date | result | age | |
|---|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative | 25 |
| 1 | Bob | 2023-08-10 | Positive | 32 |
| 2 | Charlie | NaN | NaN | 45 |
Hopefully you’re getting the hang of this and could predict that output! Since people was the right dataframe, and we are using a right join, all the rows from people are kept—Alice, Bob, and Charlie—but only matching records from test_info_xavier.
The graphic below illustrates this process:

An important point—the same final dataframe can be created with either a left join or a right join; it just depends on what order you provide the dataframes to these functions:
# Here, right join prioritizes the right dataframe, people
pd.merge(test_info_xavier, people, on='name', how='right')| name | test_date | result | age | |
|---|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative | 25 |
| 1 | Bob | 2023-08-10 | Positive | 32 |
| 2 | Charlie | NaN | NaN | 45 |
# Here, left join prioritizes the left dataframe, again people
pd.merge(people, test_info_xavier, on='name', how='left')| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
| 2 | Charlie | 45 | NaN | NaN |
As we previously mentioned, data scientists typically favor left joins over right joins. It makes more sense to specify your primary dataset first, in the left position. Opting for a left join is a common best practice due to its clearer logic, making it less error-prone.
21.14 inner join
What makes an inner join distinct is that rows are only kept if the joining values are present in both dataframes. Let’s return to our example of individuals and their test results. As a reminder, here are our datasets:
people| name | age | |
|---|---|---|
| 0 | Alice | 25 |
| 1 | Bob | 32 |
| 2 | Charlie | 45 |
test_info_xavier| name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 2023-08-10 | Positive |
| 2 | Xavier | 2023-05-02 | Negative |
Now that we have a better understanding of how joins work, we can already picture what the final dataframe would look like if we used an inner join on our two dataframes above. If only rows with joining values that are in both dataframes are kept, and the only individuals that are in both people and test_info_xavier are Alice and Bob, then they should be the only individuals in our final dataset! Let’s try it out.
pd.merge(people, test_info_xavier, on='name', how='inner')| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
Perfect, that’s exactly what we expected! Here, Charlie was only in the people dataset, and Xavier was only in the test_info_xavier dataset, so both of them were removed. The graphic below shows how this join works:

Note that the default join type is inner. So if you don’t specify how='inner', you’re actually performing an inner join! Try it out:
pd.merge(people, test_info_xavier)| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25 | 2023-06-05 | Negative |
| 1 | Bob | 32 | 2023-08-10 | Positive |
21.15 Practice Q: Inner Join Products
The following data is on product sales and customer feedback in 2019.
total_sales| product | total_units_sold | |
|---|---|---|
| 0 | Laptop | 9751 |
| 1 | Desktop | 136 |
| 2 | Tablet | 8285 |
| 3 | Smartphone | 2478 |
| 4 | Smartwatch | 3642 |
| 5 | Headphones | 5231 |
| 6 | Monitor | 1892 |
| 7 | Keyboard | 4267 |
| 8 | Mouse | 3891 |
| 9 | Printer | 982 |
product_feedback| product | n_positive_reviews | n_negative_reviews | |
|---|---|---|---|
| 0 | Laptop | 1938 | 42 |
| 1 | Desktop | 128 | 30 |
| 2 | Tablet | 842 | 56 |
| 3 | Smartphone | 1567 | 89 |
| 4 | Smartwatch | 723 | 34 |
| 5 | Headphones | 956 | 28 |
| 6 | Monitor | 445 | 15 |
| 7 | Gaming Console | 582 | 11 |
| 8 | Camera | 234 | 8 |
| 9 | Speaker | 678 | 25 |
Use an inner join to combine the datasets.
How many products are there in common between the two datasets.
Which product has the highest ratio of positive reviews to units sold? (Should be desktops)
21.16 outer join
The peculiarity of the outer join is that it retains all records, regardless of whether or not there is a match between the two datasets. Where there is missing information in our final dataset, cells are set to NaN just as we have seen in the left and right joins. Let’s take a look at our people and test_info_xavier datasets to illustrate this.
Here is a reminder of our datasets:
people
test_info_xavier| name | test_date | result | |
|---|---|---|---|
| 0 | Alice | 2023-06-05 | Negative |
| 1 | Bob | 2023-08-10 | Positive |
| 2 | Xavier | 2023-05-02 | Negative |
Now let’s perform an outer join:
pd.merge(people, test_info_xavier, on='name', how='outer')| name | age | test_date | result | |
|---|---|---|---|---|
| 0 | Alice | 25.0 | 2023-06-05 | Negative |
| 1 | Bob | 32.0 | 2023-08-10 | Positive |
| 2 | Charlie | 45.0 | NaN | NaN |
| 3 | Xavier | NaN | 2023-05-02 | Negative |
As we can see, all rows were kept so there was no loss in information! The graphic below illustrates this process:

Just as we saw above, all of the data from both of the original dataframes are still there, with any missing information set to NaN.
21.17 Practice Q: Join Sales Data
The following dataframes contain global sales and global customer complaints from various years.
sales| year | sales_count | |
|---|---|---|
| 0 | 2010 | 69890 |
| 1 | 2011 | 66507 |
| 2 | 2014 | 59831 |
| 3 | 2016 | 58704 |
| 4 | 2017 | 59151 |
customer_complaints| year | complaints_count | |
|---|---|---|
| 0 | 2011 | 1292 |
| 1 | 2013 | 1100 |
| 2 | 2015 | 1011 |
| 3 | 2016 | 940 |
| 4 | 2019 | 895 |
Join the above tables using the appropriate join to retain all information from the two datasets.
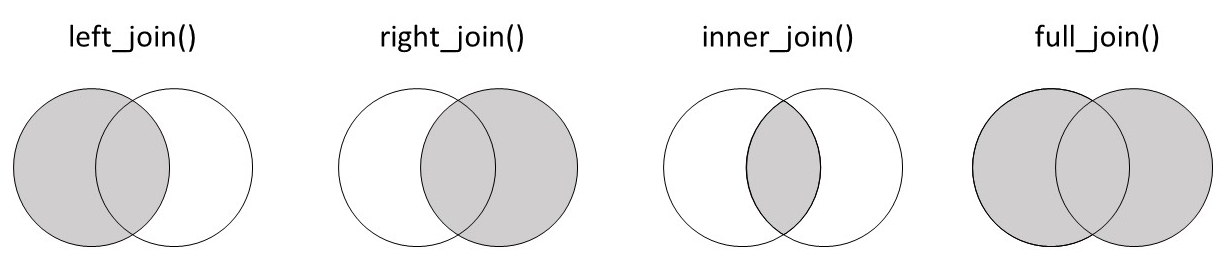
21.18 Wrap Up!
Way to go, you now understand the basics of joining! The Venn diagram below gives a helpful summary of the different joins and the information that each one retains. It may be helpful to save this image for future reference!